CellRank2教程2-快速入门
0. 概览
一般来说,基于Python的较好的工具包的一篇Notebook教程内容比较丰富,图比较多,如果直接运行跑好多张结果图,会晕乎地不知道逻辑。因此我推荐先把Notebook里的所有图粘贴出来,按照章节编好号,先通过图理解他的整个故事链之后,再开始动手运行。我们这一系列后续教程都会先带大家理解整条故事链,再去实现代码。
序言与数据预处理
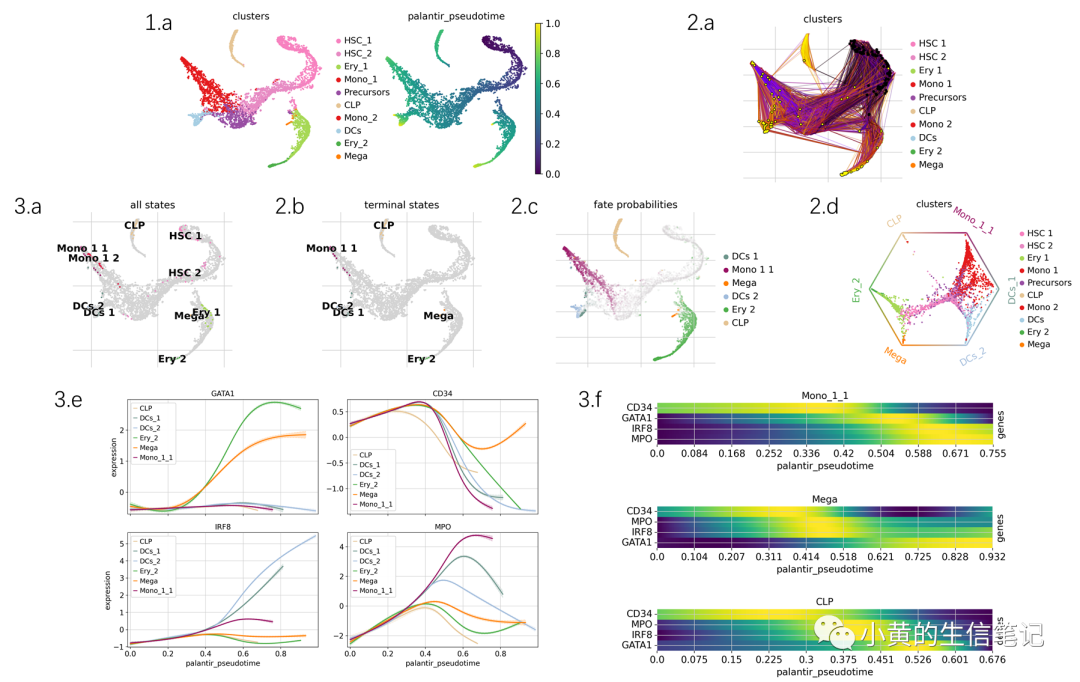
(a) 人类骨髓数据集,分别按照细胞类型着色,Palantir估计的伪时间着色。这里造血干细胞HSC出现在早期,而Mono1,CLP,DCs出现在晚期,已经很贴合生物背景了。来看看CellRank能在此基础上做些什么?
使用Kernel
(a) 指定起点(黑色),在使用kernel计算出的转移矩阵上随机游走,计算出游走的终点为Mono1, DCs,Ery1(黄色),这是转移矩阵的一种表现形式。
使用estimators (a) GPCCA在转移概率矩阵聚类出宏状态(指定状态为10个恰好与细胞类型数目相等,与细胞类型并不是完全对应的,比如DCs分为Dcs1,这里绘图下划线_有点问题)。这里的宏状态是另一种表现形式。 (b)只展示终止状态。 (c)到达所有终止状态的命运概率,颜色表示了目标,透明度表示概率。 (d)每个点代表了细胞,颜色标识细胞类型,终止状态排列在多边形的顶点,靠近哪个顶点就是越有可能往那个终止状态分化,多边形中心的细胞认为是分化程度比较低的细胞。 (e)沿着伪时间变化,各种宏状态的基因表达变化折线图,不同线对应着不同的宏状态。 (f)沿着伪时间变化,特定宏状态下各种基因的表达热图,不同行对应着不同的基因。
1. 序言与数据预处理
1.1 导入包与数据
import warnings
import cellrank as cr
import scanpy as sc
warnings.simplefilter("ignore", category=UserWarning) # 过滤警告
sc.settings.set_figure_params(frameon=False, dpi=100) # 设置图片格式
cr.settings.verbosity = 2 # 设置CellRank日志输出模式
导入人类骨髓数据集
# adata = cr.datasets.bone_marrow()
# 使用迅雷下载到本地:https://figshare.com/ndownloader/files/35826944
adata = sc.read("/home/huang/PyCode/scRNA/data/BoneMarrow/setty_bone_marrow.h5ad")
adata
AnnData object with n_obs × n_vars = 5780 × 27876
obs: 'clusters', 'palantir_pseudotime', 'palantir_diff_potential'
var: 'palantir'
uns: 'clusters_colors', 'palantir_branch_probs_cell_types'
obsm: 'MAGIC_imputed_data', 'X_tsne', 'palantir_branch_probs'
layers: 'spliced', 'unspliced'
MAGIC方法插补的表达矩阵
adata = adata[:, adata.var["palantir"]].copy()
adata.layers["MAGIC_imputed_data"] = adata.obsm["MAGIC_imputed_data"].copy()
1.2 数据预处理
scanpy标准预处理:质控、归一化、取log、找高变基因
sc.pp.filter_genes(adata, min_cells=5)
sc.pp.normalize_total(adata, target_sum=1e4)
sc.pp.log1p(adata)
sc.pp.highly_variable_genes(adata, n_top_genes=3000)
scanpy标准PCA,K近邻
sc.tl.pca(adata, random_state=0)
sc.pp.neighbors(adata, random_state=0)
t-SNE降维后展示细胞类型和Palantir估计的伪时间
sc.pl.embedding(adata, basis="tsne", color=["clusters", "palantir_pseudotime"])
2. 使用Kernel
2.1 设置Kernel
cellrank.kernels包有很多kernel,此处使用Palantir估计的伪时间结果使用PseudotimeKernel
from cellrank.kernels import PseudotimeKernel
pk = PseudotimeKernel(adata, time_key="palantir_pseudotime") # 指定伪时间的obs属性
pk
PseudotimeKernel[n=5780]
计算转移矩阵
pk.compute_transition_matrix()
pk
Computing transition matrix based on pseudotime
0%| | 0/5780 [00:00<?, ?cell/s]
Finish (0:00:00)
PseudotimeKernel[n=5780, dnorm=False, scheme='hard', frac_to_keep=0.3]
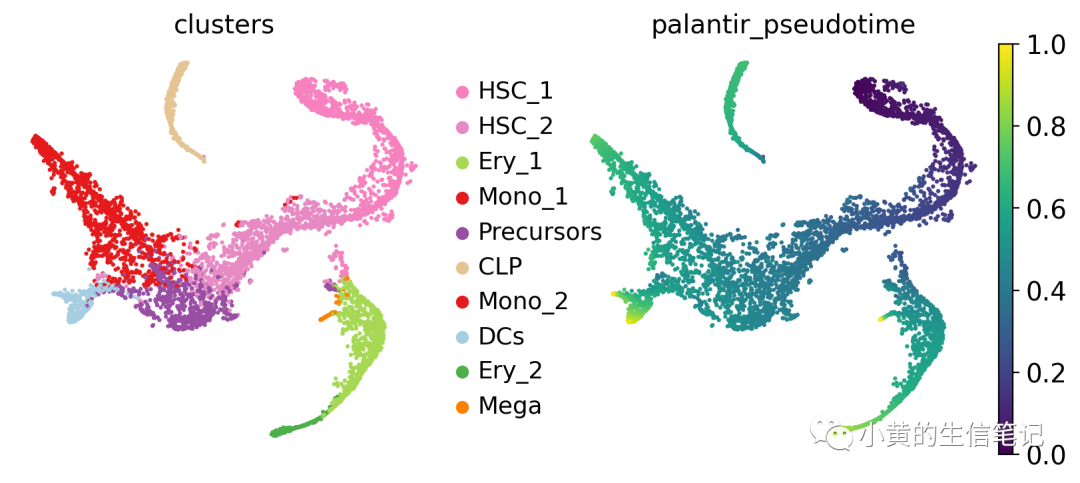
指定起点HSCs,模拟随机游走,在t-SNE降维上可视化
pk.plot_random_walks(
seed=0,
n_sims=100,
start_ixs={"clusters": "HSC_1"},
basis="tsne",
legend_loc="right",
dpi=150,
)
Simulating `100` random walks of maximum length `1445`
0%| | 0/100 [00:00<?, ?sim/s]
Finish (0:00:08)
Plotting random walks
黑色和红色分表表示随机游走的起点和终点,反映了人类造血系统中的层级分化关系
3. 使用estimators
3.1 设置estimator
Estimator使用kernel的结果并分析它,把状态空间解码成多个宏状态代表动态过程,初始、终止、中间状态。CellRank目前有两个estimator类在cellrank.estimators:
CFLARE: 聚类并过滤左右特征向量,基于转移矩阵谱的启发式方法。 GPCCA: 广义Perron聚类分析,Galerkin投影来最大化转移概率。
这里使用CFLARE
from cellrank.estimators import GPCCA
g = GPCCA(pk_new)
g
GPCCA[kernel=PseudotimeKernel[n=5780], initial_states=None, terminal_states=None]
3.2 鉴定初始、终止状态
estimator进行拟合来计算宏状态,包括初始、终止与中间状态。
g.fit(n_states=10, cluster_key="clusters")
g.plot_macrostates(which="all")
WARNING: Unable to import `petsc4py` or `slepc4py`. Using `method='brandts'`
WARNING: For `method='brandts'`, dense matrix is required. Densifying
Computing Schur decomposition
Adding `adata.uns['eigendecomposition_fwd']`
`.schur_vectors`
`.schur_matrix`
`.eigendecomposition`
Finish (0:00:45)
Computing `10` macrostates
WARNING: Color sequence contains non-unique elements
Adding `.macrostates`
`.macrostates_memberships`
`.coarse_T`
`.coarse_initial_distribution
`.coarse_stationary_distribution`
`.schur_vectors`
`.schur_matrix`
`.eigendecomposition`
Finish (0:00:08)
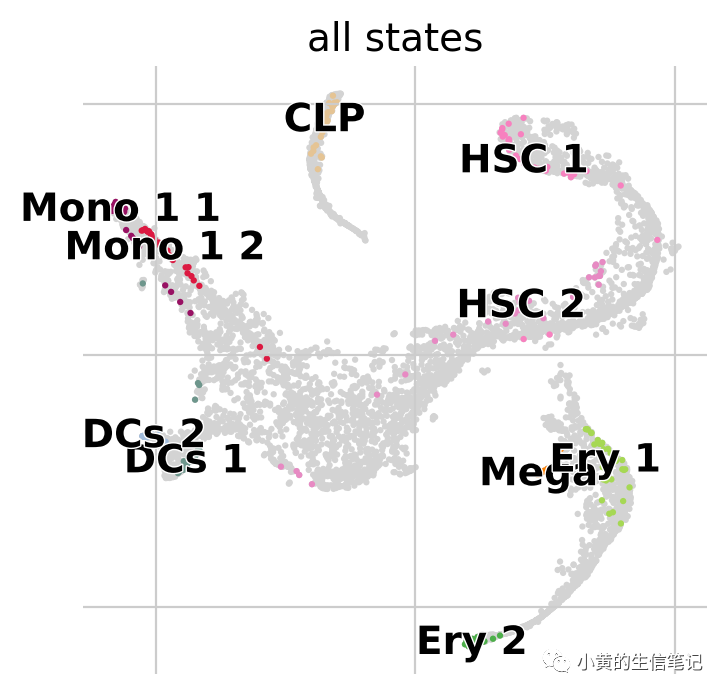
对每个宏状态,算法都会计算相关的稳定值,使用最稳定的状态
g.predict_terminal_states(method="top_n", n_states=6)
g.plot_macrostates(which="terminal")
Adding `adata.obs['term_states_fwd']`
`adata.obs['term_states_fwd_probs']`
`.terminal_states`
`.terminal_states_probabilities`
`.terminal_states_memberships
Finish`
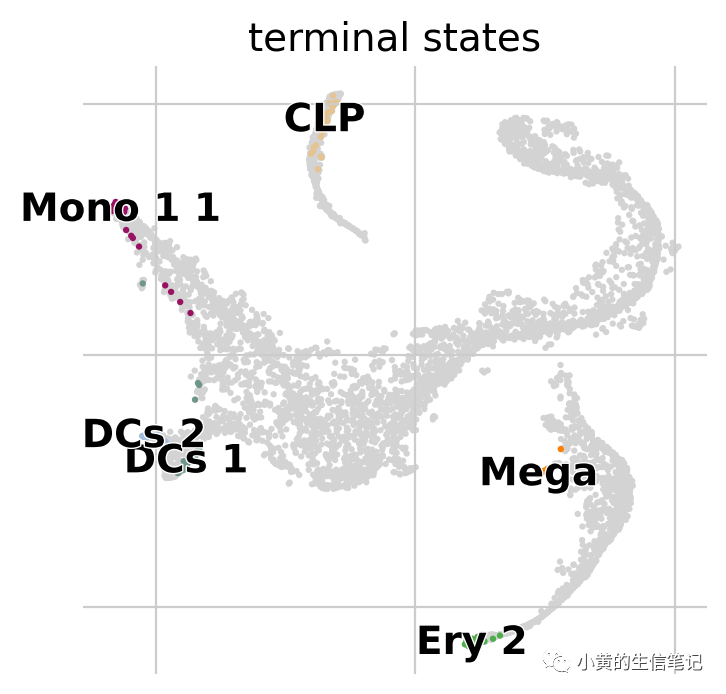
在这个数据集上,正好反映了所有的终止状态。初始状态可以使用Palantir的root很好地确定
3.3 计算命运概率和驱动基因
对每个细胞计算它到达终止状态的概率。
g.compute_fate_probabilities()
g.plot_fate_probabilities(legend_loc="right")
Computing fate probabilities
WARNING: Unable to import petsc4py. For installation, please refer to: https://petsc4py.readthedocs.io/en/stable/install.html.
Defaulting to `'gmres'` solver.
0%| | 0/6 [00:00<?, ?/s]
Adding `adata.obsm['lineages_fwd']`
`.fate_probabilities`
Finish (0:00:00)
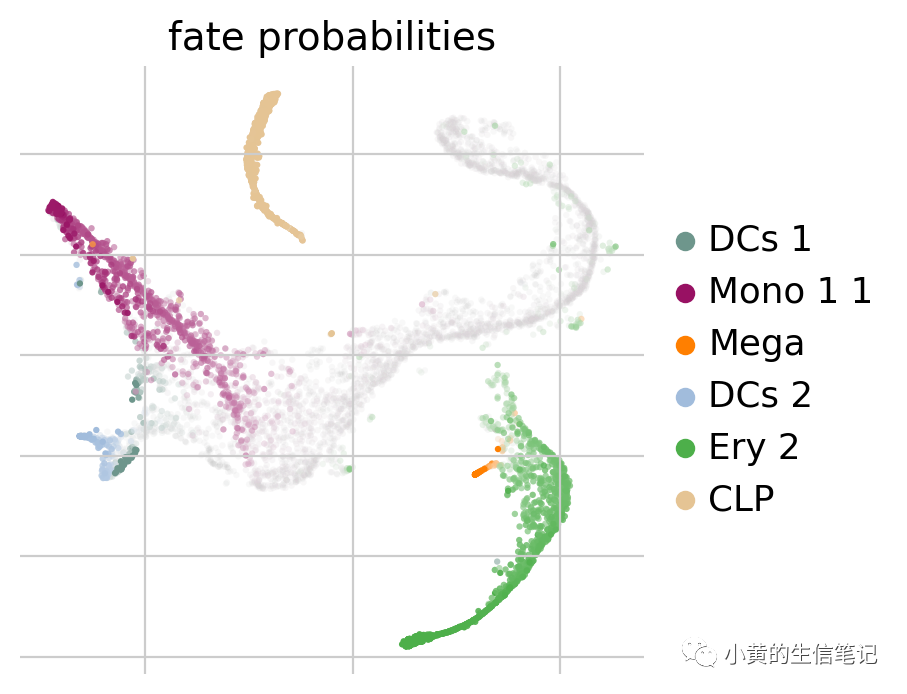
这幅图展示了到达所有终止状态的命运概率,颜色表示了目标,透明度表示概率。上述可以看到原本的大部分HSCs会往Mono11分化,也可以使用下列的环图进行绘制(这里TSP是个旅行商问题)。
cr.pl.circular_projection(adata, keys="clusters", legend_loc="right")
Solving TSP for `6` states
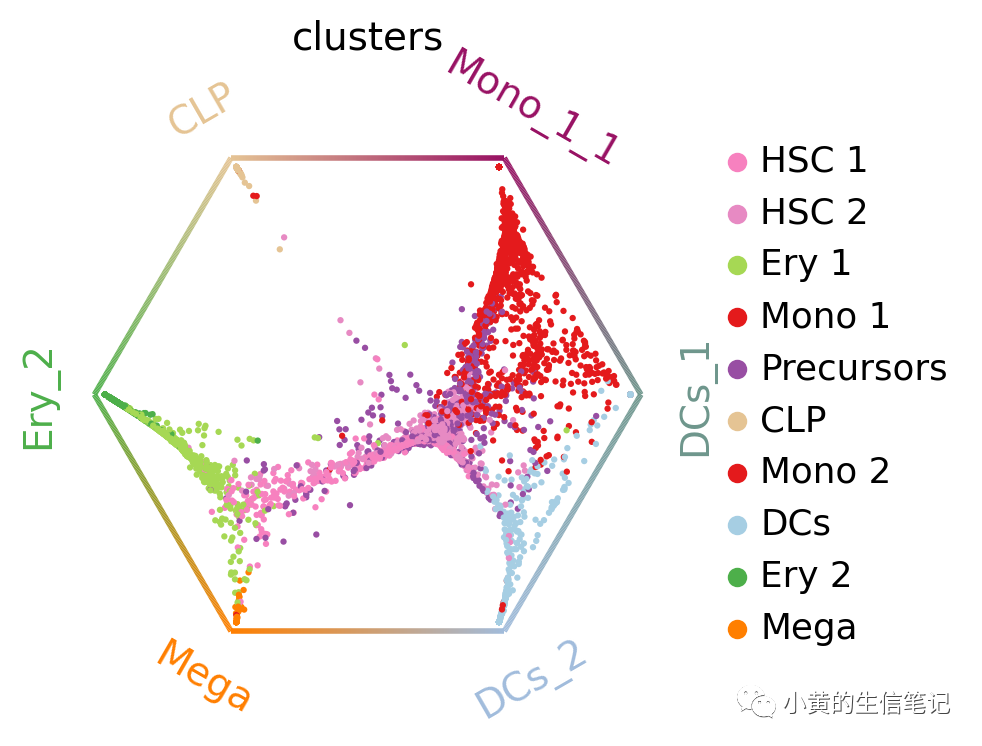
每个点代表了细胞,颜色标识细胞类型,终止状态排列在多边形的顶点,靠近哪个顶点就是越有可能往那个终止状态分化,多边形中心的细胞认为是分化程度比较低的细胞。可以看到大部分HSC倾向于往Mono_1_1分化
推断这些轨迹的驱动基因,关联表达值与命运概率,下面以Mono_1_1为例
mono_drivers = g.compute_lineage_drivers(lineages="Mono_1_1")
mono_drivers.head(10)
Adding `adata.varm['terminal_lineage_drivers']`
`.lineage_drivers`
Finish (0:00:00)
| Mono_1_1_corr | Mono_1_1_pval | Mono_1_1_qval | Mono_1_1_ci_low | Mono_1_1_ci_high | |
|---|---|---|---|---|---|
| index | |||||
| AZU1 | 0.774134 | 0.0 | 0.0 | 0.763592 | 0.784262 |
| MPO | 0.741929 | 0.0 | 0.0 | 0.730113 | 0.753301 |
| ELANE | 0.736222 | 0.0 | 0.0 | 0.724186 | 0.747809 |
| CTSG | 0.721925 | 0.0 | 0.0 | 0.709346 | 0.734044 |
| PRTN3 | 0.721322 | 0.0 | 0.0 | 0.708720 | 0.733463 |
| CFD | 0.614436 | 0.0 | 0.0 | 0.598130 | 0.630233 |
| RNASE2 | 0.595186 | 0.0 | 0.0 | 0.578278 | 0.611583 |
| MS4A3 | 0.581171 | 0.0 | 0.0 | 0.563838 | 0.597992 |
| SRGN | 0.570445 | 0.0 | 0.0 | 0.552793 | 0.587584 |
| CSTA | 0.559386 | 0.0 | 0.0 | 0.541413 | 0.576848 |
3.4 可视化基因表达趋势
给定命运概率和伪时间,可以绘制基因表达趋势
model = cr.models.GAMR(adata)
cr.pl.gene_trends(
adata,
model=model,
data_key="MAGIC_imputed_data",
genes=["GATA1", "CD34", "IRF8", "MPO"],
same_plot=True,
ncols=2,
time_key="palantir_pseudotime",
hide_cells=True,
)
Computing trends using `1` core(s)
0%| | 0/4 [00:00<?, ?gene/s]
Finish (0:00:01)
Plotting trends
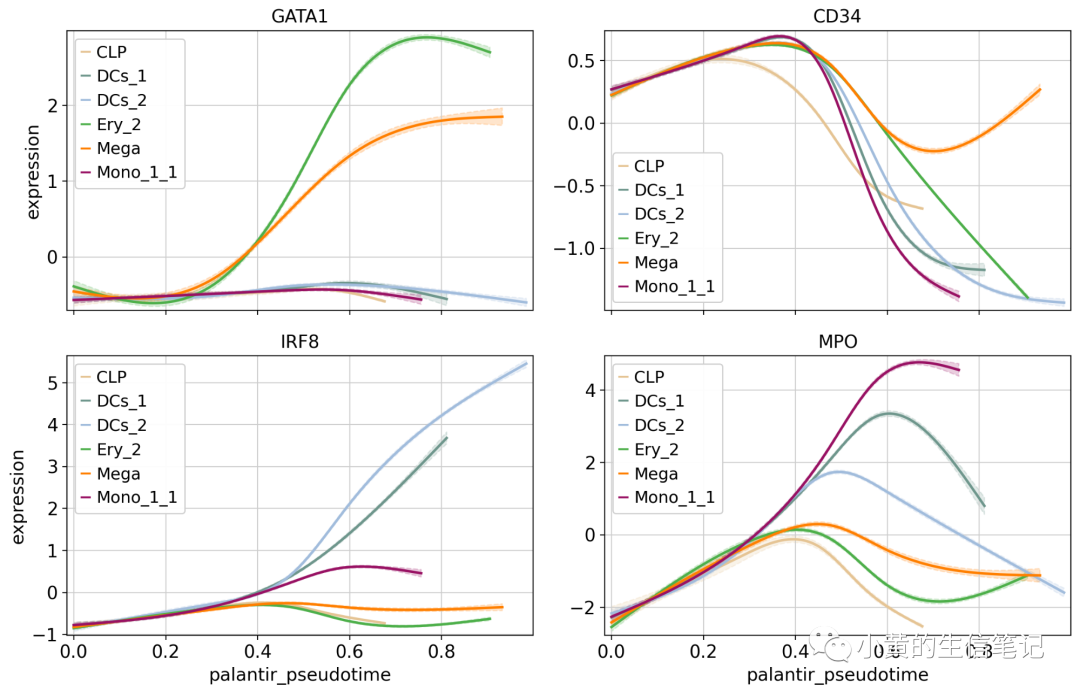
使用MAGIC插补的数据进行基因趋势的可视化,但不用于其他各项任务。然后使用热图查看多个基因变化的趋势。
cr.pl.heatmap(
adata,
model=model,
data_key="MAGIC_imputed_data",
genes=["GATA1", "CD34", "IRF8", "MPO"],
lineages=["Mono_1_1", "Mega", "CLP"],
time_key="palantir_pseudotime",
cbar=False,
show_all_genes=True,
)
Computing trends using `1` core(s)
0%| | 0/4 [00:00<?, ?gene/s]
Finish (0:00:00)
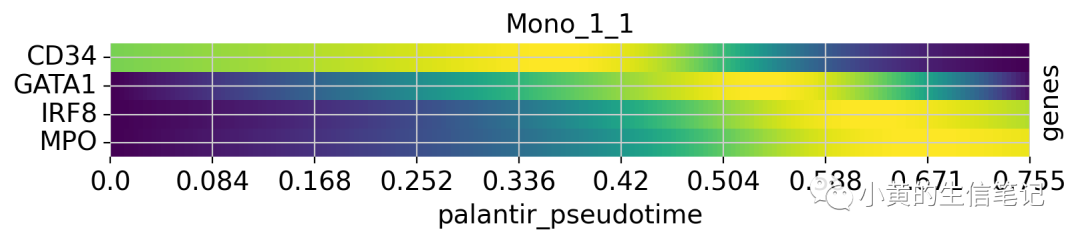
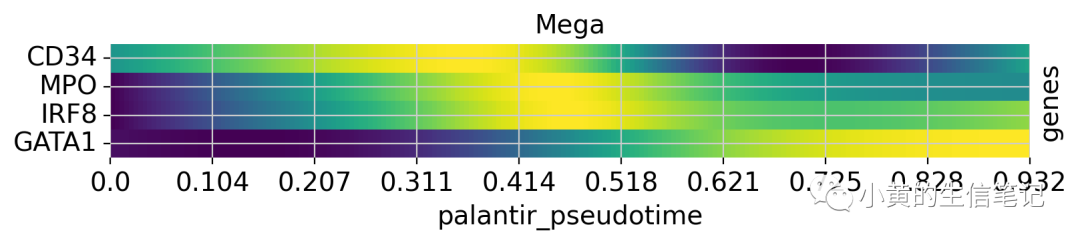
4. 接下来
接下来的教程:
探索各种CellRank的kernel和对应教程。 查看Pseudotime的文档来学习参数含义,适配自己的数据。 estimator相关:深挖关于初始、终止状态、命运概率、基因趋势的教程。