#100 CT重建的3D深度学习技术——60篇深度学习重建算法文章总结

日更100天
Lotus周六,周日,周一更新放射学前沿
全文字数6753字,阅读需要3min。
CT是快速发展的最重要的医学影像诊断手段之一。
2023年发表在Tomography的文章通过广泛的文献调查,研究三维深度学习在计算机断层扫描重建中的应用。
具体而言回答了以下2问题:
当前计算机断层扫描重建中的3D深度学习有哪些先进方法?
有哪些数据集可用于训练和验证计算机断层扫描重建中的三维深度学习?
Lotus将会为各位读者带来含有Lotus查阅了大量资料的,
关于这篇CT重建算法的综述的系列文献解读:
Part160篇3D深度学习CT重建文献
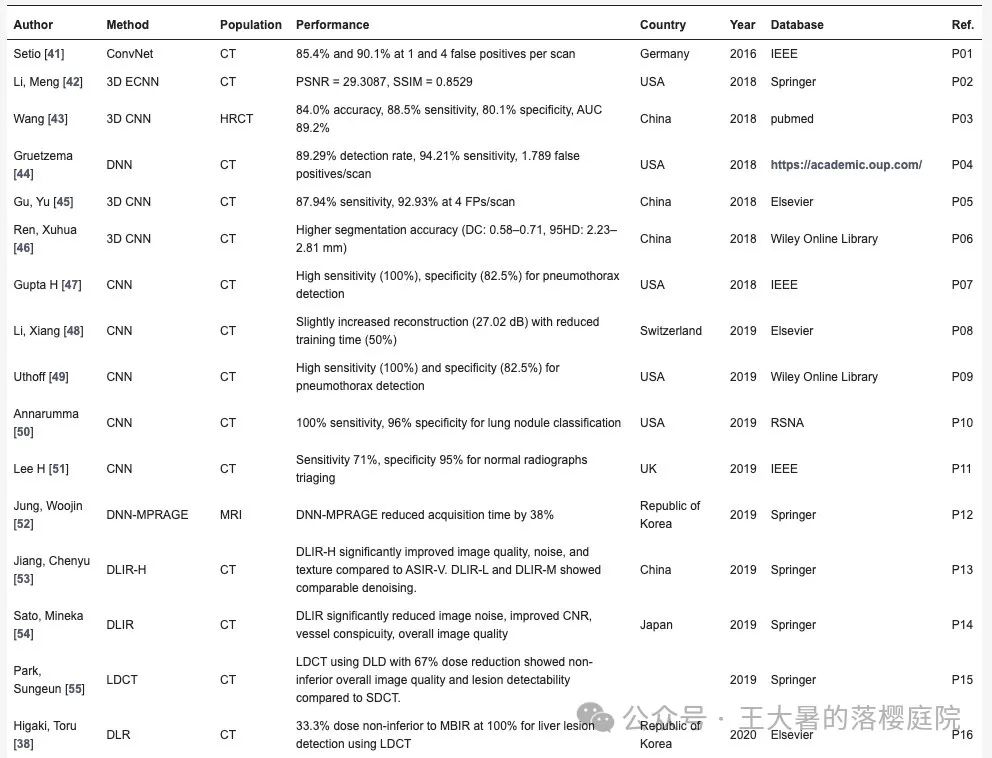
我们今天来把原文中摘选的60条经典参考文献以及主要结论进行翻译
在翻译的过程中,Lotus发现原文有一些问题,均进行了调整。
原文列出来的文章当中,有2篇重复,实际上表格列的只有58篇。
重复的两篇分别是
原文的P27和P31,使用的均为本推送中的Ref27
原文的P30和P58,使用的均为本推送中的Ref32
所以Lotus去掉了原文的第32行(P31)和第59行(P58),
将这两行与原文的第28行和第30行进行了合并。
注明:
最后的ref后面的数字对应本推送末尾的参考文献,
如果有感兴趣的朋友可以对应去相应数据库进行进一步的阅读。
| 作者 | 方法 | 人群 | 表现 | 国家 | 年 | 数据库 | ref |
|---|---|---|---|---|---|---|---|
| Setio | ConvNet | CT | 每次扫描出现1个和4个假阳性率分别为 85.4%和90.1% | 德国 | 2016 | IEEE | R1 |
| Li, Meng | 3D ECNN | CT | PSNR = 29.3087, SSIM = 0.8529 | 美国 | 2018 | Springer | R2 |
| Wang | 3D CNN | HRCT | 84.0% accuracy, 88.5% sensitivity, 80.1% specificity, AUC 89.2% | 中国 | 2018 | pubmed | R3 |
| Gruetzema | DNN | CT | 检测率(detection rate)89.29% , 94.21% sensitivity, 每次扫描出现1.789个假阳性 | 美国 | 2018 | academic.oup | R4 |
| Gu, Yu | 3D CNN | CT | 87.94% sensitivity, 每次扫描出现4个假阳性率为92.93% | 中国 | 2018 | Elsevier | R5 |
| Ren, Xuhua | 3D CNN | CT | 更高的分割正确率(DC: 0.58–0.71, 95HD: 2.23–2.81 mm) | 中国 | 2018 | Wiley | R6 |
| Gupta H | CNN | CT | 气胸检测灵敏度(sensitivity)高(100%),特异性(specificity)强(82.5%)。 | 美国 | 2018 | IEEE | R7 |
| Li, Xiang | CNN | CT | 在重建27.02dB的重建数据上可以减少50%的训练时间 | 瑞士 | 2019 | Elsevier | R8 |
| Uthoff | CNN | CT | 【Lotus:觉得这篇应该不符合标准,这是一篇组学模型文章】肺结节识别模型100% sensitivity,96% specificity | 瑞士 | 2019 | Elsevier | R9 |
| Annarumma | CNN | X光 | sensitivity 71%, specificity 95%, PPV 73%, and NPV 94% | 瑞士 | 2019 | RSNA | R10 |
| Lee H | DLIR | CT | 降低67%辐射剂量实现诊断非劣效性 | 韩国 | 2019 | Springer | R11 |
| Jung, Woojin | DNN-MPRAGE | MRI | 减少38%的重建时间 | 韩国 | 2022 | Springer | R12 |
| Jiang, Chenyu | DLIR | CT | 与 ASIR-V 相比,DLIR-H 明显改善了图像质量、噪声和纹理。DLIR-L 和 DLIR-M 的去噪效果相当。 | 中国 | 2022 | Springer | R13 |
| Sato, Mineka | DLIR | CT | DLIR 大大降低了图像噪声,提高了 CNR、血管清晰度和整体图像质量 | 日本 | 2022 | Springer | R14 |
| Park, Sungeun | DLIR | CT | 与MBIR相比,在肝脏诊断上降低67%辐射剂量实现诊断非劣效性 | 韩国 | 2022 | Springer | R15 |
| Higaki, Toru | DLR | CT | 与MBIR相比,在肝脏诊断上降低67%辐射剂量实现诊断非劣效性 | 日本 | 2020 | MDPI | R16 |
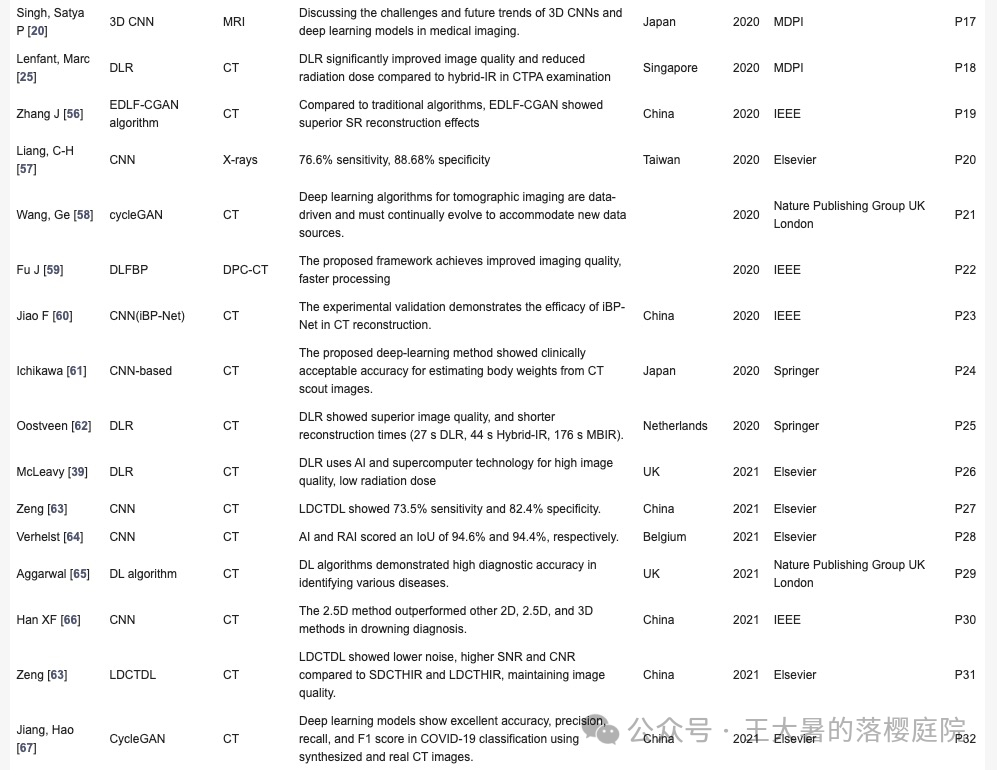
| Singh, Satya P | 3D CNN | MRI | 讨论3D CNN和深度学习模型在医学成像中的挑战和未来趋势。 | 日本 | 2020 | MDPI | R17 |
| Lenfant, Marc | 3D CNN | CT | 青光眼和健康眼在内部和外部验证集上为99.6%和91.0% | 日本 | 2019 | MDPI | R18 |
| Zhang J | EDLF-CGAN | CT | 与传统算法相比,EDLF-CGAN 显示出卓越的 SR 重建效果 | 中国 | 2020 | IEEE | R19 |
| Liang, C-H | CNN | X线 | 76.6% sensitivity, 88.68% specificity | 中国 | 2020 | Elsevier | R20 |
| Wang, Ge | cycleGAN | CT | 断层成像的深度学习算法是数据驱动的,必须不断发展以适应新的数据源。 | 美国 | 2020 | Nature | R21 |
| Fu J | DLFBP | CT | 建议的框架可提高成像质量,加快处理 | 中国 | 2020 | IEEE | R22 |
| Jiao F | iBP-Net | CT | 实验验证了iBP-Net的CT重建效率 | 中国 | 2020 | IEEE | R23 |
| Ichikawa | CNN | CT | 深度学习方法在从 CT 扫描图像估算体重方面显示出了临床上可接受的准确性。 | 中国 | 2020 | Springer | R24 |
| Oostveen | DLR | CT | DLR 显示出卓越的图像质量和更短的重建时间 | 荷兰 | 2020 | Springer | R25 |
| McLeavy | DL | CT | 人工智能和超级计算机技术实现高图像质量和低辐射剂量 | 荷兰 | 2021 | Elsevier | R26 |
| Zeng | CNN | CT | 与 SDCTHIR 和 LDCTHIR 相比,LDCTDL 显示出更低的噪声、更高的 SNR 和 CNR,同时保持了图像质量。73.5% sensitivity和 82.4% specificity | 中国 | 2021 | Elsevier | R27 |
| Verhelst | CNN | CT | AI 和 RAI 的 IoU 分别为 94.6% 和 94.4%。 | 比利时 | 2021 | Elsevier | R28 |
| Aggarwal | DL | CT | DL 算法在识别各种疾病方面具有很高的诊断准确性。 | 英国 | 2021 | Nature | R29 |
| Han XF | CNN | CT | 所开发的深度学习网络能够对三维骨骼模型进行高精度估算。 | 中国 | 2021 | IEEE | R30 |
| Jiang, Hao | CycleGAN | CT | 在使用合成和真实 CT 图像进行COVID-19分类时,深度学习模型显示出出色的accuracy, precision, recall和F1分数。 | 中国 | 2021 | Elsevier | R31 |
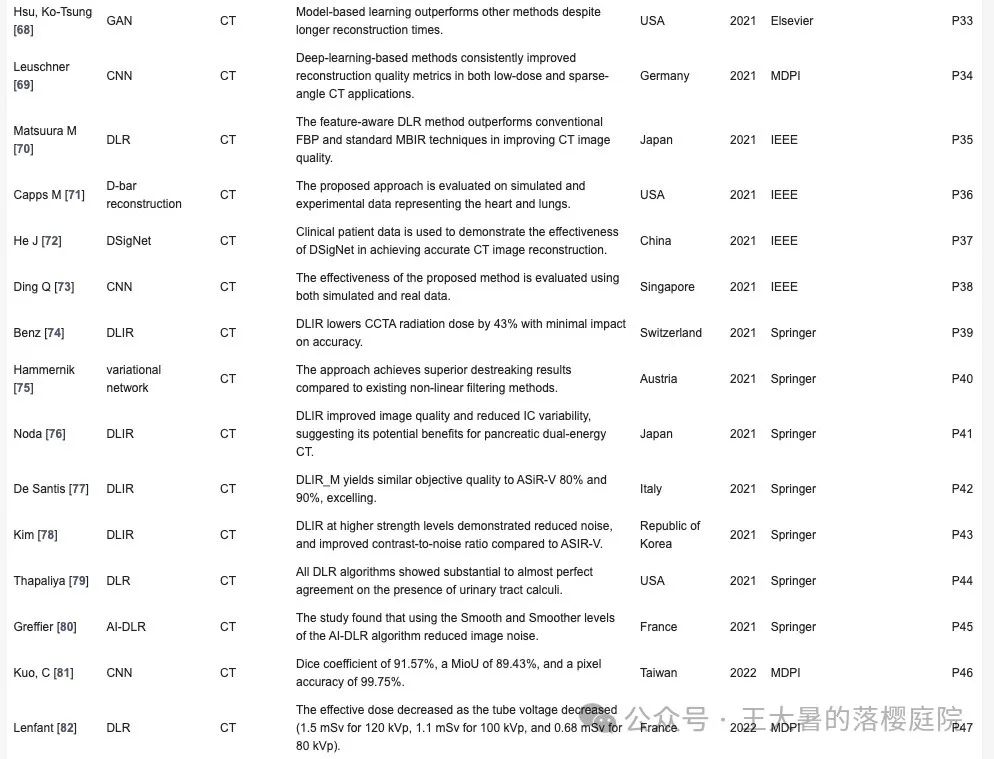
| Hsu, Ko-Tsung | GAN | CT | 尽管重建时间较长,但基于模型的学习方法优于其他方法。 | 美国 | 2021 | Elsevier | R32 |
| Leuschner | CNN | CT | 在低剂量和稀疏角度CT应用中,基于深度学习的方法持续改善了重建质量指标。 | 德国 | 2021 | MDPI | R33 |
| Matsuura M | CNN | CT | 在提高 CT 图像质量方面,特征感知DL方法优于传统的FBP和标准MBIR技术。 | 日本 | 2020 | IEEE | R34 |
| Capps M | D-bar | CT | 所提出的方法在代表心脏和肺部的模拟和实验数据上进行了评估。 | 美国 | 2020 | IEEE | R35 |
| He J | DSigNet | CT | 临床患者数据用于证明DSigNet在实现精确CT图像重建方面的有效性。 | 中国 | 2021 | IEEE | R36 |
| Ding Q | CNN | CT | 我们利用模拟数据和真实数据对所提方法的有效性进行了评估。 | 新加坡 | 2021 | IEEE | R37 |
| Benz | DLIR | CT | DLIR可将CCTA辐射剂量降低43%,而对准确性的影响却微乎其微。 | 瑞士 | 2022 | Springer | R38 |
| Hammernik | DL | CT | 与现有的非线性滤波方法相比,该方法取得了更优越的去除应力效果。 | 澳大利亚 | 2017 | Springer | R39 |
| Noda | DLIR | CT | DLIR提高了图像质量,降低了IC测量差异,这表明它对胰腺双能CT 有潜在的益处。 | 日本 | 2022 | Springer | R40 |
| De Santis | DLIR | CT | DLIR_M的客观质量与ASiR-V80%和90%相比,更胜一筹。 | 意大利 | 2023 | Springer | R41 |
| Kim | DLIR | CT | 与 ASIR-V 相比,DLIR 在更高强度水平上降低了噪声,提高了对比度-噪声比。 | 韩国 | 2021 | Springer | R42 |
| Thapaliya | DL | CT | 所有DL算法在尿路结石诊断一致性良好。 | 美国 | 2022 | Springer | R43 |
| Greffier | DL | CT | AI重建算法降低图像噪声 | 法国 | 2023 | Springer | R44 |
| Kuo, C | CNN | CT | Dice coefficient 91.57%,MioU 为 89.43%,像素精度(pixel accuracy)为 99.75%。 | 中国 | 2022 | MDPI | R45 |
| Lenfant | DLR | CT | 有效剂量随着电子管电压的降低而降低(120 kVp为 1.5 mSv,100 kVp为1.1 mSv,80 kVp为0.68 mSv)。 | 法国 | 2022 | MDPI | R46 |
| Hu D | DEAR | CBCT | 使用从商用扫描仪获取的锥形束乳腺CT数据集对DEAR网络的性能进行了评估。 | 美国 | 2020 | IEEE | R47 |
| Xie H | PWLS | CT | 我们利用十名患者的临床 SDCT和模拟LDCT扫描结果,证明了所提出方法的有效性。 | 美国 | 2022 | IEEE | R48 |
| Park HS | wGAN | CT | 机器学习方法wGAN的图像质量优于FBP。 | 澳大利亚 | 2020 | Springer | R49 |
| Thaler | U-Net | CT | U-Net性能最佳,MAE 最低,PSNR 最高,SSIM 最高。 | 日本 | 2018 | Springer | R50 |
| Koike | DLIR | CT | 与hybrid-IR相比,DLIR的使用大大降低了图像噪声,提高了胰腺 LDCT图像的质量。 | 日本 | 2022 | Springer | R51 |
| Noda | DLR | CT | 就图像噪声而言,在肝动脉期和平衡期,LD DLR和LD MBIR图像优于MBIR SD hybrid-IR图像。 | 日本 | 2022 | Springer | R52 |
| Nakamura | DLIR | CT | DLIR 以小于 50% 的辐射剂量获得了与上腹部胸部 CT 相当的图像质量。 | 韩国 | 2021 | Springer | R53 |
| Nam | DLIR | CT | 在上腹部扫描中,与迭代重建直接比较,DLIR图像质量相似,辐射剂量更低。 | 韩国 | 2021 | Springer | R54 |
| Shin | 3D DPI | CT | 成功观测完整小鼠肺呼气时的三维肺泡单元并测量肺泡直径。 | 韩国 | 2021 | Nature | R55 |
| Zeng Y | VAE | CT | VAE模型的灵敏度(sensitivity)为 79.2%,特异度(specificity)为 72.7%,准确度(accuracy)为 77.1%,F1分数为 0.667,AUROC 为 0.801。 | 韩国 | 2023 | Nature | R56 |
| Shiode | CNN | X线 | 3D bone model能够有效的从腕关节的X光片中重建得到3D的腕关节图像。 | 日本 | 2021 | Nature | R57 |
| Bornet | DLR | CT | 与MBIR图像相比,DLR的图像噪明显更低,CNR也更高。 | 法国 | 2022 | Springer | R58 |
Part2原文
(未完待续)
参考文献
Setio, A.A.A.; Ciompi, F.; Litjens, G.; Gerke, P.; Jacobs, C.; Van Riel, S.J.; Wille, M.M.W.; Naqibullah, M.; Sánchez, C.I.; Van Ginneken, B. Pulmonary nodule detection in CT images: False positive reduction using multi-view convolutional networks. IEEE Trans. Med. Imaging 2016, 35, 1160–1169. [Google Scholar] [CrossRef] Li, M.; Shen, S.; Gao, W.; Hsu, W.; Cong, J. Computed tomography image enhancement using 3D convolutional neural network. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 291–299. [Google Scholar] Wang, S.; Wang, R.; Zhang, S.; Li, R.; Fu, Y.; Sun, X.; Li, Y.; Sun, X.; Jiang, X.; Guo, X.; et al. 3D convolutional neural network for differentiating pre-invasive lesions from invasive adenocarcinomas appearing as ground-glass nodules with diameters ≤ 3 cm using HRCT. Quant. Imaging Med. Surg. 2018, 8, 491. [Google Scholar] [CrossRef] Gruetzemacher, R.; Gupta, A.; Paradice, D. 3D deep learning for detecting pulmonary nodules in CT scans. J. Am. Med. Informatics Assoc. 2018, 25, 1301–1310. [Google Scholar] [CrossRef] Gu, Y.; Lu, X.; Yang, L.; Zhang, B.; Yu, D.; Zhao, Y.; Gao, L.; Wu, L.; Zhou, T. Automatic lung nodule detection using a 3D deep convolutional neural network combined with a multi-scale prediction strategy in chest CTs. Comput. Biol. Med. 2018, 103, 220–231. [Google Scholar] [CrossRef] Ren, X.; Xiang, L.; Nie, D.; Shao, Y.; Zhang, H.; Shen, D.; Wang, Q. Interleaved 3D-CNN s for joint segmentation of small-volume structures in head and neck CT images. Med. Phys. 2018, 45, 2063–2075. [Google Scholar] [CrossRef] Gupta, H.; Jin, K.H.; Nguyen, H.Q.; McCann, M.T.; Unser, M. CNN-based projected gradient descent for consistent CT image reconstruction. IEEE Trans. Med. Imaging 2018, 37, 1440–1453. [Google Scholar] [CrossRef] [PubMed] Li, X.; Thrall, J.H.; Digumarthy, S.R.; Kalra, M.K.; Pandharipande, P.V.; Zhang, B.; Nitiwarangkul, C.; Singh, R.; Khera, R.D.; Li, Q. Deep learning-enabled system for rapid pneumothorax screening on chest CT. Eur. J. Radiol. 2019, 120, 108692. [Google Scholar] [CrossRef] [PubMed] Uthoff, J.; Stephens, M.J.; Newell, J.D., Jr.; Hoffman, E.A.; Larson, J.; Koehn, N.; De Stefano, F.A.; Lusk, C.M.; Wenzlaff, A.S.; Watza, D.; et al. Machine learning approach for distinguishing malignant and benign lung nodules utilizing standardized perinodular parenchymal features from CT. Med. Phys. 2019, 46, 3207–3216. [Google Scholar] [CrossRef] [PubMed] Annarumma, M.; Withey, S.J.; Bakewell, R.J.; Pesce, E.; Goh, V.; Montana, G. Automated triaging of adult chest radiographs with deep artificial neural networks. Radiology 2019, 291, 196–202. [Google Scholar] [CrossRef] [PubMed] Lee, H.; Lee, J.; Kim, H.; Cho, B.; Cho, S. Deep-neural-network-based sinogram synthesis for sparse-view CT image reconstruction. IEEE Trans. Radiat. Plasma Med. Sci. 2018, 3, 109–119. [Google Scholar] [CrossRef] Jung, W.; Kim, J.; Ko, J.; Jeong, G.; Kim, H.G. Highly accelerated 3D MPRAGE using deep neural network–based reconstruction for brain imaging in children and young adults. Eur. Radiol. 2022, 32, 5468–5479. [Google Scholar] [CrossRef] [PubMed] Jiang, C.; Jin, D.; Liu, Z.; Zhang, Y.; Ni, M.; Yuan, H. Deep learning image reconstruction algorithm for carotid dual-energy computed tomography angiography: Evaluation of image quality and diagnostic performance. Insights Imaging 2022, 13, 182. [Google Scholar] [CrossRef] Sato, M.; Ichikawa, Y.; Domae, K.; Yoshikawa, K.; Kanii, Y.; Yamazaki, A.; Nagasawa, N.; Nagata, M.; Ishida, M.; Sakuma, H. Deep learning image reconstruction for improving image quality of contrast-enhanced dual-energy CT in abdomen. Eur. Radiol. 2022, 32, 5499–5507. [Google Scholar] [CrossRef] Park, S.; Yoon, J.H.; Joo, I.; Yu, M.H.; Kim, J.H.; Park, J.; Kim, S.W.; Han, S.; Ahn, C.; Kim, J.H.; et al. Image quality in liver CT: Low-dose deep learning vs standard-dose model-based iterative reconstructions. Eur. Radiol. 2022, 32, 2865–2874. [Google Scholar] [CrossRef] Higaki, T.; Nakamura, Y.; Zhou, J.; Yu, Z.; Nemoto, T.; Tatsugami, F.; Awai, K. Deep learning reconstruction at CT: Phantom study of the image characteristics. Acad. Radiol. 2020, 27, 82–87. [Google Scholar] [CrossRef] [PubMed] Singh, S.P.; Wang, L.; Gupta, S.; Goli, H.; Padmanabhan, P.; Gulyás, B. 3D deep learning on medical images: A review. Sensors 2020, 20, 5097. [Google Scholar] [CrossRef] MacCormick, I.J.; Williams, B.M.; Zheng, Y.; Li, K.; Al-Bander, B.; Czanner, S.; Cheeseman, R.; Willoughby, C.E.; Brown, E.N.; Spaeth, G.L.; et al. Accurate, fast, data efficient and interpretable glaucoma diagnosis with automated spatial analysis of the whole cup to disc profile. PLoS ONE 2019, 14, e0209409. [Google Scholar] Zhang, J.; Gong, L.R.; Yu, K.; Qi, X.; Wen, Z.; Hua, Q.; Myint, S.H. 3D reconstruction for super-resolution CT images in the Internet of health things using deep learning. IEEE Access 2020, 8, 121513–121525. [Google Scholar] [CrossRef] Liang, C.H.; Liu, Y.C.; Wu, M.T.; Garcia-Castro, F.; Alberich-Bayarri, A.; Wu, F.Z. Identifying pulmonary nodules or masses on chest radiography using deep learning: External validation and strategies to improve clinical practice. Clin. Radiol. 2020, 75, 38–45. [Google Scholar] [CrossRef] [PubMed] Wang, G.; Ye, J.C.; De Man, B. Deep learning for tomographic image reconstruction. Nat. Mach. Intell. 2020, 2, 737–748. [Google Scholar] [CrossRef] Fu, J.; Dong, J.; Zhao, F. A deep learning reconstruction framework for differential phase-contrast computed tomography with incomplete data. IEEE Trans. Image Process. 2019, 29, 2190–2202. [Google Scholar] [CrossRef] [PubMed] Jiao, F.; Gui, Z.; Li, K.; Shangguang, H.; Wang, Y.; Liu, Y.; Zhang, P. A dual-domain CNN-based network for CT reconstruction. IEEE Access 2021, 9, 71091–71103. [Google Scholar] [CrossRef] Ichikawa, S.; Hamada, M.; Sugimori, H. A deep-learning method using computed tomography scout images for estimating patient body weight. Sci. Rep. 2021, 11, 15627. [Google Scholar] [CrossRef] Oostveen, L.J.; Meijer, F.J.; de Lange, F.; Smit, E.J.; Pegge, S.A.; Steens, S.C.; van Amerongen, M.J.; Prokop, M.; Sechopoulos, I. Deep learning–based reconstruction may improve non-contrast cerebral CT imaging compared to other current reconstruction algorithms. Eur. Radiol. 2021, 31, 5498–5506. [Google Scholar] [CrossRef] McLeavy, C.; Chunara, M.; Gravell, R.; Rauf, A.; Cushnie, A.; Talbot, C.S.; Hawkins, R. The future of CT: Deep learning reconstruction. Clin. Radiol. 2021, 76, 407–415. [Google Scholar] [CrossRef] Zeng, L.; Xu, X.; Zeng, W.; Peng, W.; Zhang, J.; Sixian, H.; Liu, K.; Xia, C.; Li, Z. Deep learning trained algorithm maintains the quality of half-dose contrast-enhanced liver computed tomography images: Comparison with hybrid iterative reconstruction: Study for the application of deep learning noise reduction technology in low dose. Eur. J. Radiol. 2021, 135, 109487. [Google Scholar] [CrossRef] Verhelst, P.J.; Smolders, A.; Beznik, T.; Meewis, J.; Vandemeulebroucke, A.; Shaheen, E.; Van Gerven, A.; Willems, H.; Politis, C.; Jacobs, R. Layered deep learning for automatic mandibular segmentation in cone-beam computed tomography. J. Dent. 2021, 114, 103786. [Google Scholar] [CrossRef] Aggarwal, R.; Sounderajah, V.; Martin, G.; Ting, D.S.; Karthikesalingam, A.; King, D.; Ashrafian, H.; Darzi, A. Diagnostic accuracy of deep learning in medical imaging: A systematic review and meta-analysis. NPJ Digit. Med. 2021, 4, 65. [Google Scholar] [CrossRef] Chung, Y.W.; Choi, I.Y. Detection of abnormal extraocular muscles in small datasets of computed tomography images using a three-dimensional variational autoencoder. Sci. Rep. 2023, 13, 1765. [Google Scholar] [CrossRef] Jiang, H.; Tang, S.; Liu, W.; Zhang, Y. Deep learning for COVID-19 chest CT (computed tomography) image analysis: A lesson from lung cancer. Comput. Struct. Biotechnol. J. 2021, 19, 1391–1399. [Google Scholar] [CrossRef] [PubMed] Hsu, K.T.; Guan, S.; Chitnis, P.V. Comparing deep learning frameworks for photoacoustic tomography image reconstruction. Photoacoustics 2021, 23, 100271. [Google Scholar] [CrossRef] [PubMed] Leuschner, J.; Schmidt, M.; Ganguly, P.S.; Andriiashen, V.; Coban, S.B.; Denker, A.; Bauer, D.; Hadjifaradji, A.; Batenburg, K.J.; Maass, P.; et al. Quantitative comparison of deep learning-based image reconstruction methods for low-dose and sparse-angle CT applications. J. Imaging 2021, 7, 44. [Google Scholar] [CrossRef] [PubMed] Matsuura, M.; Zhou, J.; Akino, N.; Yu, Z. Feature-aware deep-learning reconstruction for context-sensitive X-ray computed tomography. IEEE Trans. Radiat. Plasma Med. Sci. 2020, 5, 99–107. [Google Scholar] [CrossRef] Capps, M.; Mueller, J.L. Reconstruction of organ boundaries with deep learning in the D-bar method for electrical impedance tomography. IEEE Trans. Biomed. Eng. 2020, 68, 826–833. [Google Scholar] [CrossRef] [PubMed] He, J.; Chen, S.; Zhang, H.; Tao, X.; Lin, W.; Zhang, S.; Zeng, D.; Ma, J. Downsampled imaging geometric modeling for accurate CT reconstruction via deep learning. IEEE Trans. Med. Imaging 2021, 40, 2976–2985. [Google Scholar] [CrossRef] [PubMed] Ding, Q.; Nan, Y.; Gao, H.; Ji, H. Deep learning with adaptive hyper-parameters for low-dose CT image reconstruction. IEEE Trans. Comput. Imaging 2021, 7, 648–660. [Google Scholar] [CrossRef] Benz, D.C.; Ersözlü, S.; Mojon, F.L.; Messerli, M.; Mitulla, A.K.; Ciancone, D.; Kenkel, D.; Schaab, J.A.; Gebhard, C.; Pazhenkottil, A.P.; et al. Radiation dose reduction with deep-learning image reconstruction for coronary computed tomography angiography. Eur. Radiol. 2022, 32, 2620–2628. [Google Scholar] [CrossRef] Hammernik, K.; Würfl, T.; Pock, T.; Maier, A. A deep learning architecture for limited-angle computed tomography reconstruction. In Proceedings of the Bildverarbeitung für die Medizin 2017: Algorithmen-Systeme-Anwendungen. Proceedings des Workshops vom 12. bis 14. März 2017 in Heidelberg; Springer: Berlin/Heidelberg, Germany, 2017; pp. 92–97. [Google Scholar] Noda, Y.; Kawai, N.; Nagata, S.; Nakamura, F.; Mori, T.; Miyoshi, T.; Suzuki, R.; Kitahara, F.; Kato, H.; Hyodo, F.; et al. Deep learning image reconstruction algorithm for pancreatic protocol dual-energy computed tomography: Image quality and quantification of iodine concentration. Eur. Radiol. 2022, 32, 384–394. [Google Scholar] [CrossRef] De Santis, D.; Polidori, T.; Tremamunno, G.; Rucci, C.; Piccinni, G.; Zerunian, M.; Pugliese, L.; Del Gaudio, A.; Guido, G.; Barbato, L.; et al. Deep learning image reconstruction algorithm: Impact on image quality in coronary computed tomography angiography. La Radiol. Medica 2023, 128, 434–444. [Google Scholar] [CrossRef] Kim, I.; Kang, H.; Yoon, H.J.; Chung, B.M.; Shin, N.Y. Deep learning–based image reconstruction for brain CT: Improved image quality compared with adaptive statistical iterative reconstruction-Veo (ASIR-V). Neuroradiology 2021, 63, 905–912. [Google Scholar] [CrossRef] [PubMed] Thapaliya, S.; Brady, S.L.; Somasundaram, E.; Anton, C.G.; Coley, B.D.; Towbin, A.J.; Zhang, B.; Dillman, J.R.; Trout, A.T. Detection of urinary tract calculi on CT images reconstructed with deep learning algorithms. Abdom. Radiol. 2022, 47, 265–271. [Google Scholar] [CrossRef] [PubMed] Greffier, J.; Durand, Q.; Frandon, J.; Si-Mohamed, S.; Loisy, M.; de Oliveira, F.; Beregi, J.P.; Dabli, D. Improved image quality and dose reduction in abdominal CT with deep-learning reconstruction algorithm: A phantom study. Eur. Radiol. 2023, 33, 699–710. [Google Scholar] [CrossRef] [PubMed] Kuo, C.F.J.; Liao, Y.S.; Barman, J.; Liu, S.C. Semi-supervised deep learning semantic segmentation for 3D volumetric computed tomographic scoring of chronic rhinosinusitis: Clinical correlations and comparison with Lund-Mackay scoring. Tomography 2022, 8, 718–729. [Google Scholar] [CrossRef] Lenfant, M.; Comby, P.O.; Guillen, K.; Galissot, F.; Haioun, K.; Thay, A.; Chevallier, O.; Ricolfi, F.; Loffroy, R. Deep Learning-Based Reconstruction vs. Iterative Reconstruction for Quality of Low-Dose Head-and-Neck CT Angiography with Different Tube-Voltage Protocols in Emergency-Department Patients. Diagnostics 2022, 12, 1287. [Google Scholar] [CrossRef] Xie, H.; Shan, H.; Wang, G. Deep encoder-decoder adversarial reconstruction (DEAR) network for 3D CT from few-view data. Bioengineering 2019, 6, 111. [Google Scholar] [CrossRef] [PubMed] Xie, H.; Shan, H.; Cong, W.; Liu, C.; Zhang, X.; Liu, S.; Ning, R.; Wang, G. Deep efficient end-to-end reconstruction (DEER) network for few-view breast CT image reconstruction. IEEE Access 2020, 8, 196633–196646. [Google Scholar] [CrossRef] Park, H.S.; Kim, K.; Jeon, K. Low-dose CT image reconstruction with a deep learning prior. IEEE Access 2020, 8, 158647–158655. [Google Scholar] [CrossRef] Thaler, F.; Hammernik, K.; Payer, C.; Urschler, M.; Štern, D. Sparse-view CT reconstruction using wasserstein GANs. In Proceedings of the International Workshop on Machine Learning for Medical Image Reconstruction, Granada, Spain, 16 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 75–82. [Google Scholar] Koike, Y.; Ohira, S.; Teraoka, Y.; Matsumi, A.; Imai, Y.; Akino, Y.; Miyazaki, M.; Nakamura, S.; Konishi, K.; Tanigawa, N.; et al. Pseudo low-energy monochromatic imaging of head and neck cancers: Deep learning image reconstruction with dual-energy CT. Int. J. Comput. Assist. Radiol. Surg. 2022, 17, 1271–1279. [Google Scholar] [CrossRef] Noda, Y.; Iritani, Y.; Kawai, N.; Miyoshi, T.; Ishihara, T.; Hyodo, F.; Matsuo, M. Deep learning image reconstruction for pancreatic low-dose computed tomography: Comparison with hybrid iterative reconstruction. Abdom. Radiol. 2021, 46, 4238–4244. [Google Scholar] [CrossRef] Nakamura, Y.; Narita, K.; Higaki, T.; Akagi, M.; Honda, Y.; Awai, K. Diagnostic value of deep learning reconstruction for radiation dose reduction at abdominal ultra-high-resolution CT. Eur. Radiol. 2021, 31, 4700–4709. [Google Scholar] [CrossRef] Nam, J.G.; Hong, J.H.; Kim, D.S.; Oh, J.; Goo, J.M. Deep learning reconstruction for contrast-enhanced CT of the upper abdomen: Similar image quality with lower radiation dose in direct comparison with iterative reconstruction. Eur. Radiol. 2021, 31, 5533–5543. [Google Scholar] [CrossRef] [PubMed] Shin, S.; Kim, M.W.; Jin, K.H.; Yi, K.M.; Kohmura, Y.; Ishikawa, T.; Je, J.H.; Park, J. Deep 3D reconstruction of synchrotron X-ray computed tomography for intact lungs. Sci. Rep. 2023, 13, 1738. [Google Scholar] [CrossRef] [PubMed] Zeng, Y.; Zhang, X.; Kawasumi, Y.; Usui, A.; Ichiji, K.; Funayama, M.; Homma, N. A 2.5 D deep learning-based method for drowning diagnosis using post-mortem computed tomography. IEEE J. Biomed. Health Informatics 2022, 27, 1026–1035. [Google Scholar] [CrossRef] [PubMed] Shiode, R.; Kabashima, M.; Hiasa, Y.; Oka, K.; Murase, T.; Sato, Y.; Otake, Y. 2D–3D reconstruction of distal forearm bone from actual X-ray images of the wrist using convolutional neural networks. Sci. Rep. 2021, 11, 15249. [Google Scholar] [CrossRef] Bornet, P.A.; Villani, N.; Gillet, R.; Germain, E.; Lombard, C.; Blum, A.; Gondim Teixeira, P.A. Clinical acceptance of deep learning reconstruction for abdominal CT imaging: Objective and subjective image quality and low-contrast detectability assessment. Eur. Radiol. 2022, 32, 3161–3172. [Google Scholar] [CrossRef]
欢迎转载和抄袭,赞赏一下,喜欢就说是你写的。
如果喜欢就点点点赞,转发,在看或者在下面留言吧。
创作不易,请Lotus喝盏茶~